Download v0.2.0
Mac and Linux builds are available on GitHub
Introduction
MMonitor is software designed for monitoring metagenomes over time, using data from nanopore sequencing. It includes a Python application for desktop use, which manages the analysis of sequence data and metadata for taxonomic and functional insight, as well as quality control. The accompanying webserver Dashboard provides a platform for users to view their analysis results after logging in. Developed with input from laboratory researchers, MMonitor supports real-time data analysis to offer immediate sample overviews, and is flexible for use on personal computers or via external servers for enhanced processing capabilities.
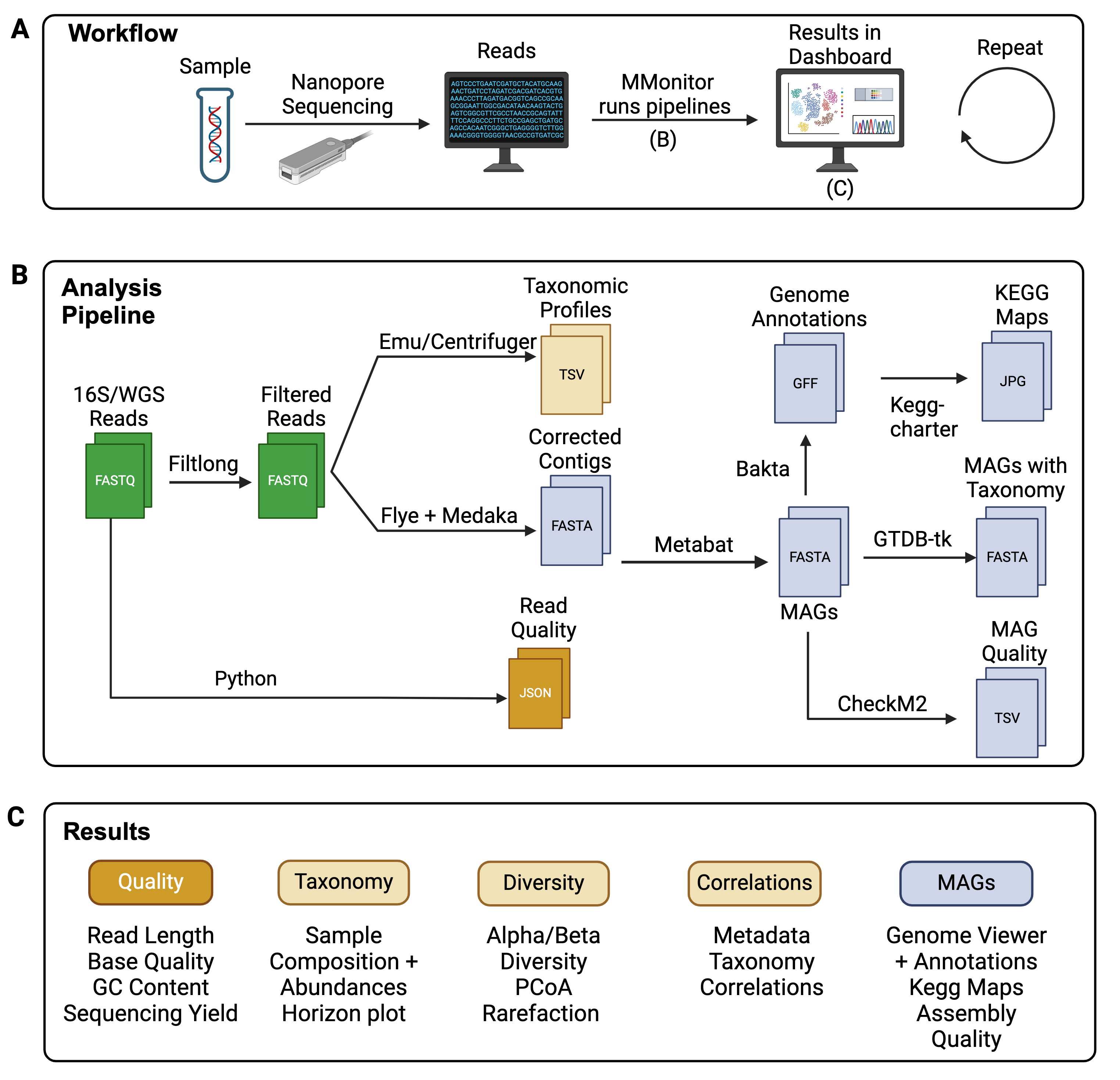
Workflow
Get started by preparing your samples and running them through nanopore sequencing. MMonitor consists of two parts: the Desktop application that you'll need to download to run the analysis pipelines, and this web Dashboard where you can view and explore your results.
Once your sequencing is complete, MMonitor Desktop will automatically process your raw data and make the results available in your personal dashboard. You can monitor the analysis progress in real-time as your data comes in.
Analysis Pipeline
MMonitor Desktop runs your sequencing data through our optimized pipeline. Your reads are first quality controlled to ensure the best results. Then, it performs two types of analysis depending on input data:
- Identify what organisms are in your sample using taxonomic profiling
- Assemble and annotate complete genomes from your data using metagenome assembly and binning
Each genome is automatically checked for quality and annotated with functional information.
Dashboard Apps
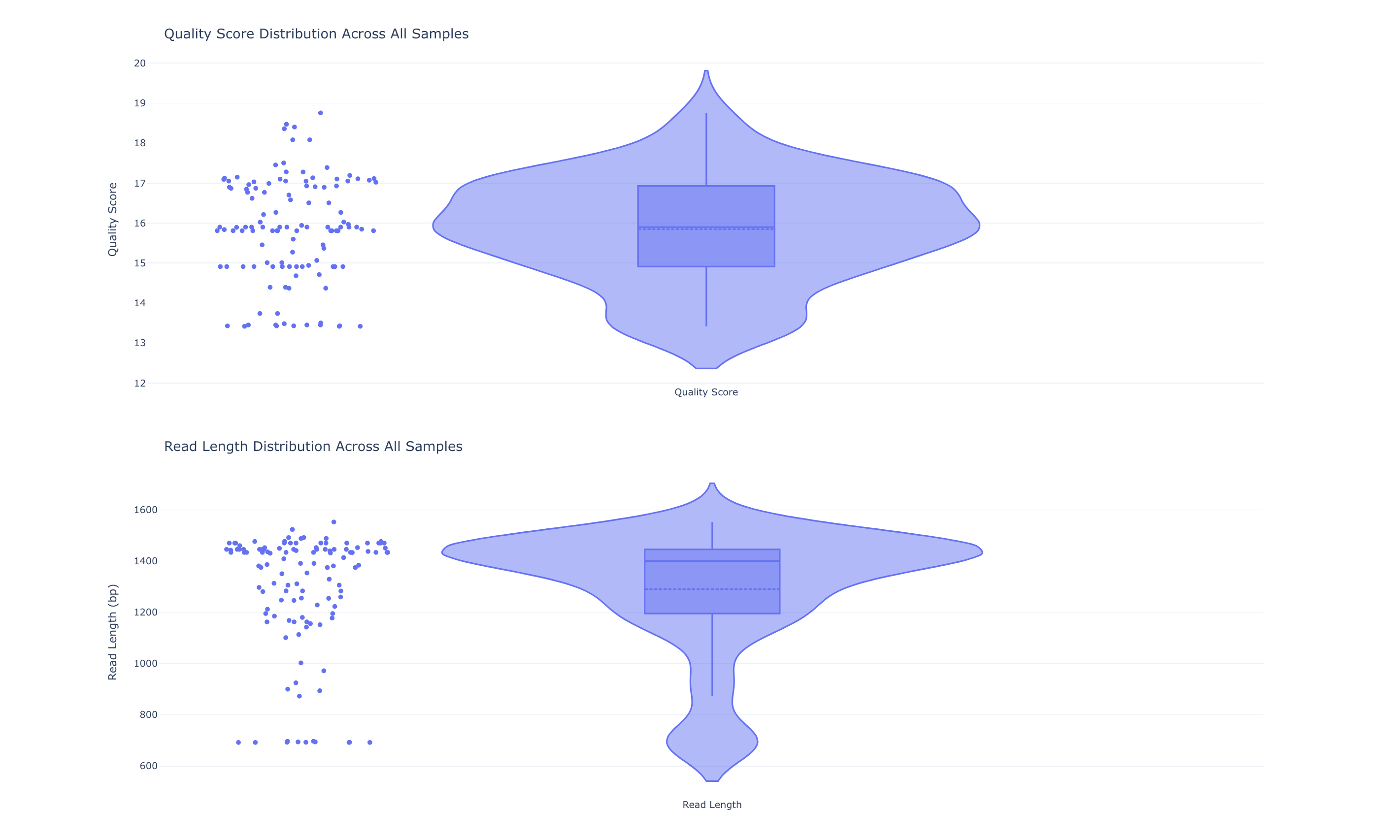
QC
Overview of common quality metrics of the input data
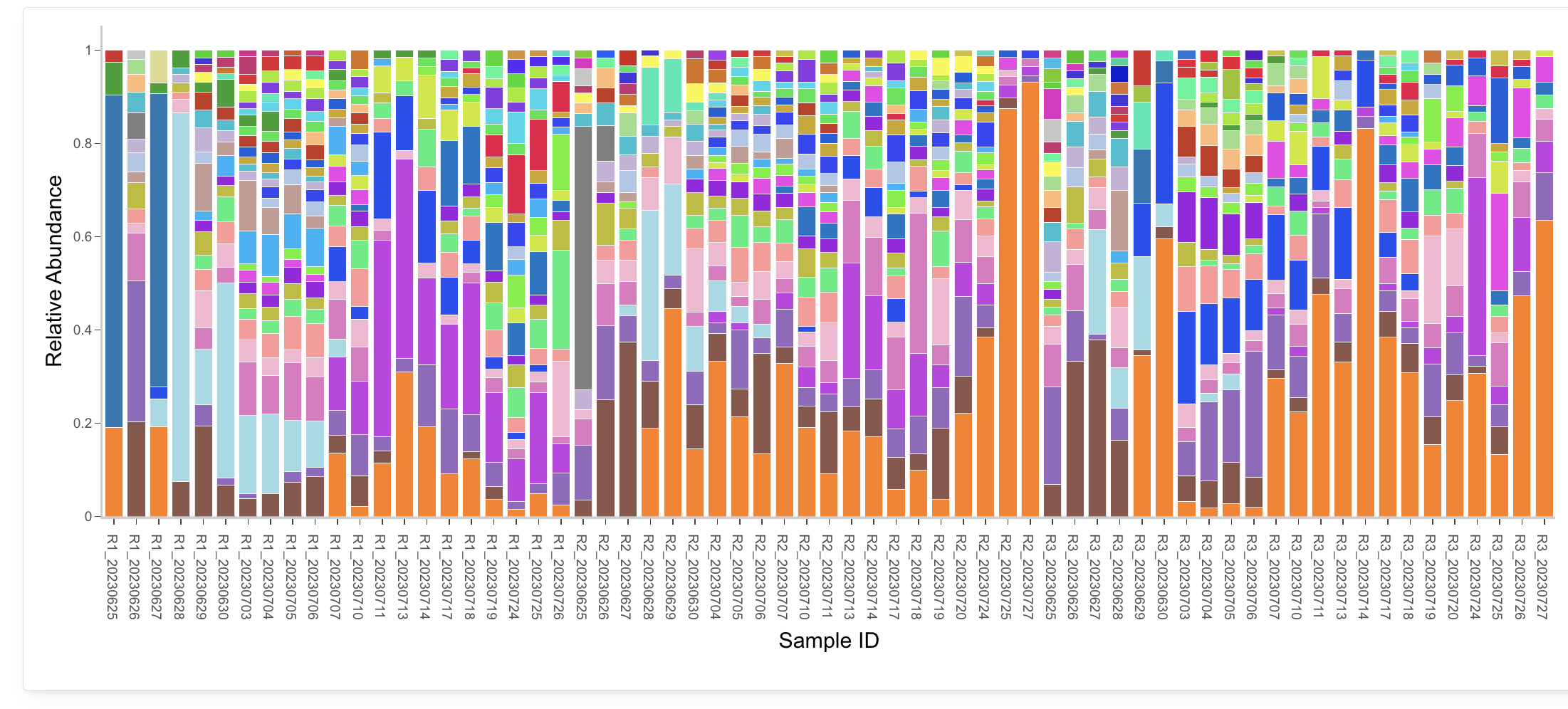
Taxonomy
Analyze the taxonomic composition of metagenome samples
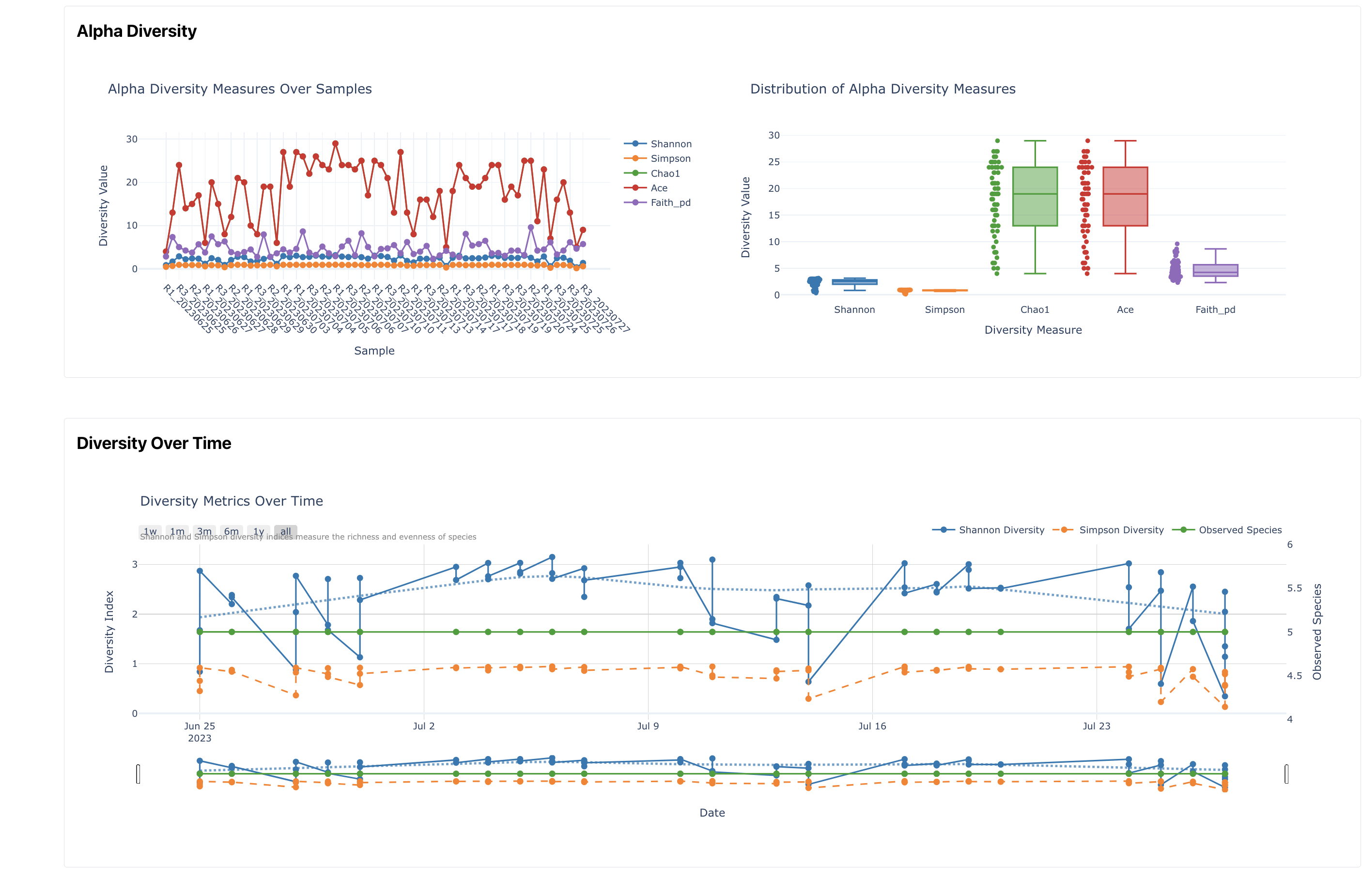
Diversity
Overview of diversity metrics, including alpha and beta diversity, PCoA plots and rarefaction curves
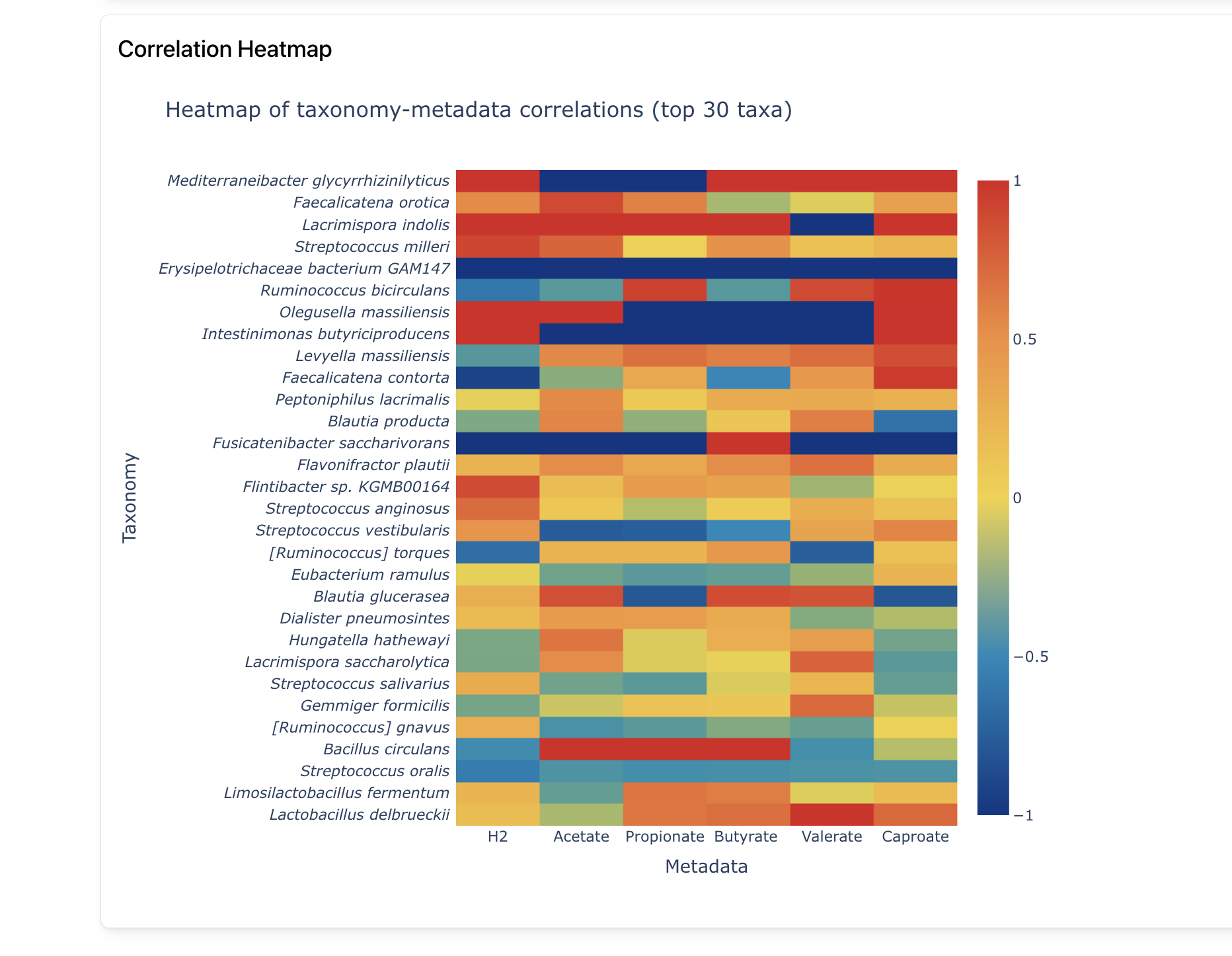
Correlations
Correlate metagenomic data with external metadata to highlight important variables
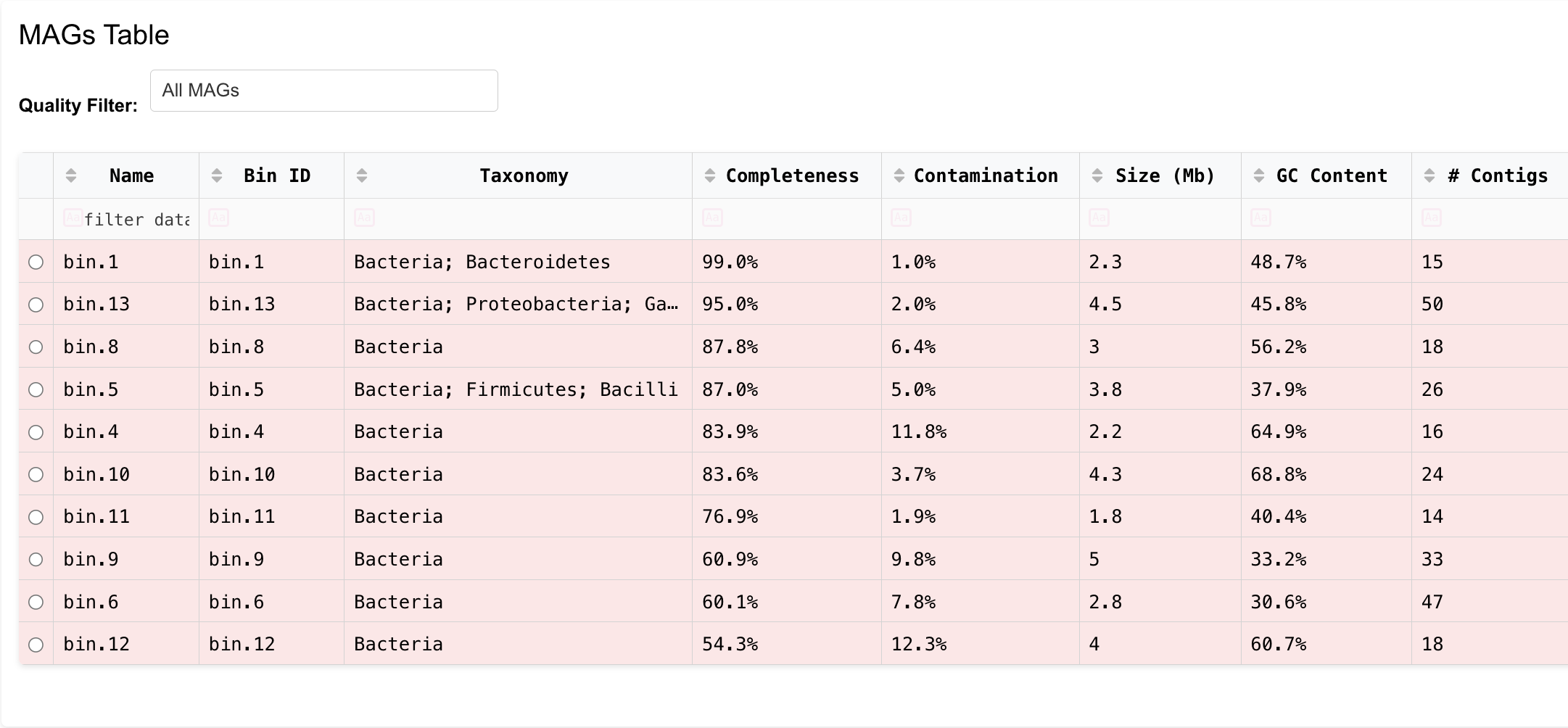
MAGs
Analyze MAGs with genome visualization and annotations
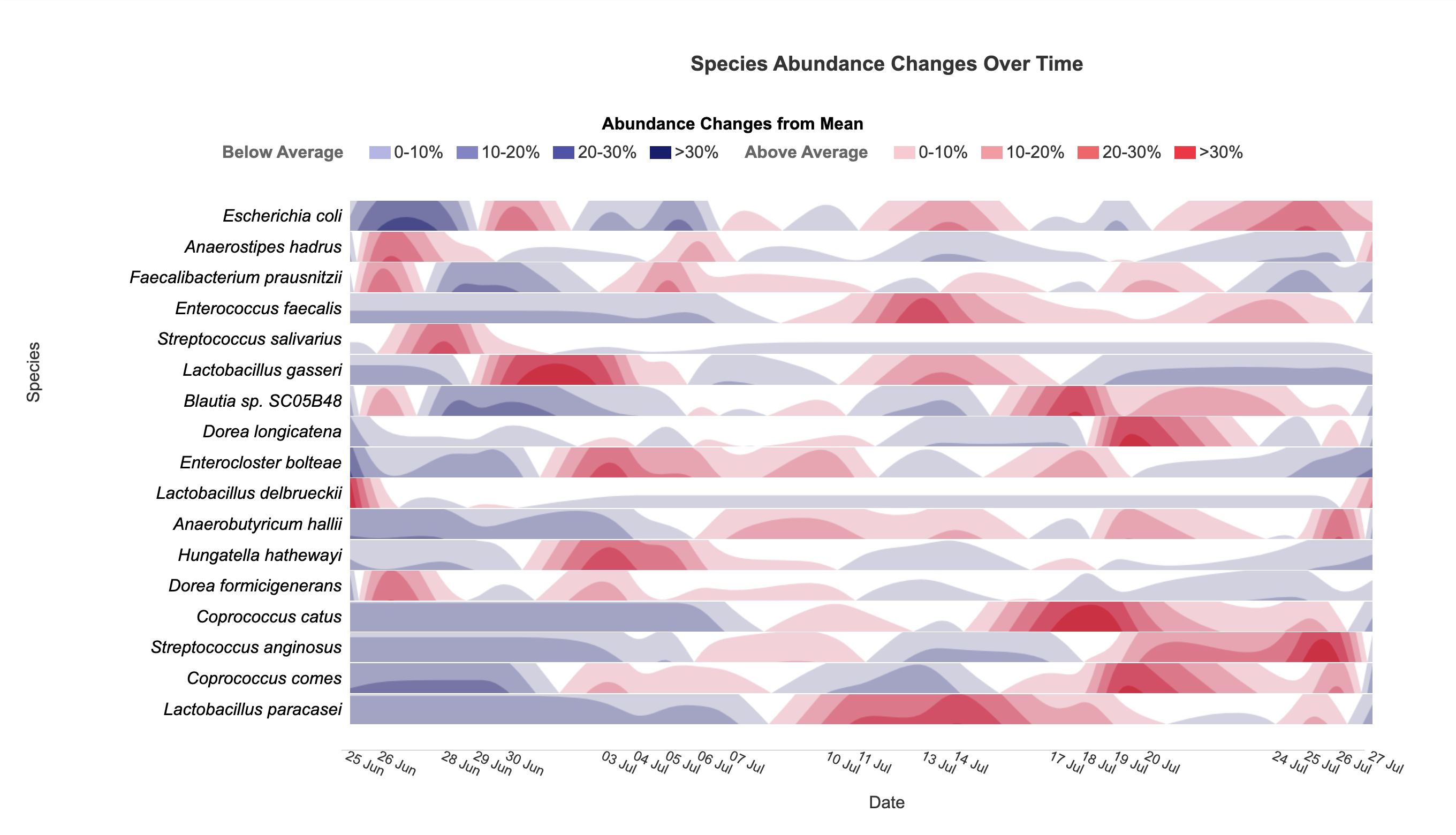
Horizon
Visualize taxonomic abundance changes over time with interactive horizon charts
Installation and Documentation
For installation instructions and detailed documentation, please visit our GitHub repository.
